University of Maryland Center for Environmental Science publishes the first full reference genome for the blue crab
University of Maryland Center for Environmental Science (UMCES) researchers at the Institute of Marine and Environmental Technology (IMET) at Baltimore’s Inner Harbor have published the first full reference genome sequence for the blue crab. This means that for the first time we have a complete picture of all of the DNA that makes up Maryland’s favorite crustacean. This information is important to science research and will contribute to a healthy crab fishery. The genome will help fisheries managers maintain sustainable blue crab stocks, leading to a healthy bay ecosystem, a strong regional economy, and many Maryland crab feasts.
“Marylanders love crabs, and everybody wants to have big, fat crabs in the fall. Understanding what makes them successful is located in the chromosomes,” said Professor Sook Chung, an expert in crab biology who led the project. “Knowing the full genome, we are several steps closer to identifying the genes responsible for growth, reproduction, and susceptibility to disease.”
The best way to understand an organism is to understand its genetic makeup, also known as its genome. The genome is the DNA sequence of the chromosomes that give the instructions for how an organism grows and develops. Once the code is understood, it reveals many secrets of how the organism works. It is akin to seeing the blueprint for a building. Once you understand the “blueprint” of an organism, you can understand what genetic traits make some crabs particularly successful at reproducing or others more adapted to changing water temperatures fueled by climate change.
Understanding not only how many crabs are in the Bay, but how likely they are to reproduce successfully could aid in fisheries policies, helping to maintain a healthy ecosystem and economy. For example, breeding particularly fertile females could help enable the production of blue crabs in aquaculture. The genome could also potentially be used for food source tracking to determine if the lump crab meat in the market came from Venezuela or Maryland’s coastal bays.
Since the genome within a species varies by individual, any genome mapping project begins with choosing the best possible sample organism. In late October 2018, Chung went out on the Chesapeake Bay on a crabber’s boat and collected dozens of young female blue crabs to breed in IMET’s Aquaculture Research Center (ARC). One female grew to adulthood, mated, and successfully produced offspring, proving she had good genes for reproducing. This crab’s daughter was selected for sequencing and dubbed “The Chosen One.”
Scientists isolated DNA from the crab and sent it off to be sequenced. The sequence of the genetic code determines how an organism will grow and develop. When the genetic code is sequenced, it is initially jumbled up from its proper order. The process of correctly ordering the code, or “assembly,” required a special computer running night and day for over six months.
“Imagine you take several volumes of an encyclopedia and you have a hundred copies of each volume. You put them all through a paper shredder and then you have to use that to reconstruct the original volumes of the encyclopedia,” said Associate Research Professor Tsvetan Bachvaroff, who was responsible for assembling the blue crab genome. “Once the encyclopedia, or genome, is back in the correct order, you can begin to identify genes and use it like a reference book, looking up genes to answer questions.”
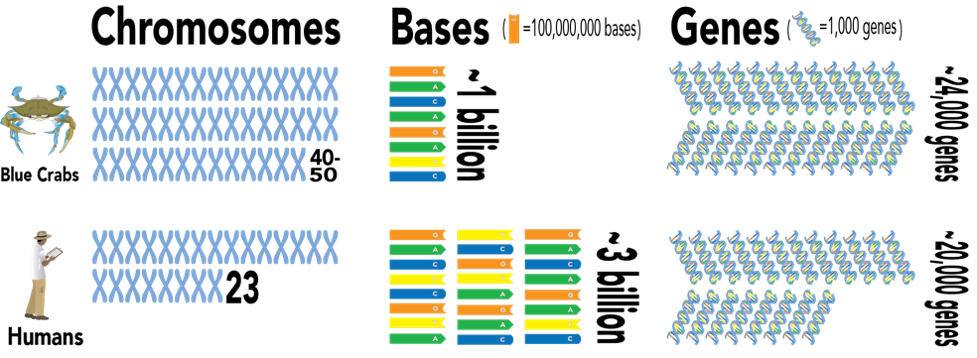
Researchers determined that the blue crab had between 40 and 50 chromosomes, which is nearly double the amount found in humans. However, these chromosomes were very short, resulting in a genome that is approximately one third the length of the human genome, in terms of bases. Despite its relatively diminutive size, the blue crab genome is rich in gene diversity, containing approximately 24,000 genes, slightly more than the amount identified in humans.
The project was funded by a novel approach, with funding provided by a small group of philanthropic Marylanders who are passionate about science, supporting IMET, the Chesapeake Bay, and the blue crab.
“Sook’s enthusiasm for this work was contagious, and we were thrilled to discover that the skills and technology for this exciting project existed right here in Baltimore,” said Mike Davis, who led the private effort to fund the project. “Her analogy of requiring a 'blueprint' for the blue crab, to really understand it, just like builders use blueprints to understand a building clearly resonated with the donors, and we all really wanted this scientific milestone achieved here in Maryland.”
The sequencing of the Blue Crab Genome was made possible by the support of the following generous donors: The G. Unger Vetlesen Foundation, Mike and Trish Davis, Don and Cathy MacMurray, James J. Albrecht, Bertram and Debbie Winchester, Arnold and Alison Richman, Maryland Sea Grant, Arthur Jib Edwards, J. Sook Chung, Richard L. Franyo, Edward St. John Foundation, Tom and Nancy Reynolds, James E. Connell, Russell T. Hill, Bill and Chris Hufnell, David Balcom, J. Mitchell Neitzey, James and Jenny Corckran, Richard and Maureen Roden, and Nicholas L. Hammond.
The paper, "Chromosome-level Genome Assembly of the Blue Crab, Callinectes sapidus," was published in G3: Genes | Genomes | Genetics by University of Maryland Center for Environmental Science scientists Tsvetan Bachvaroff, Ryan McDonald, Louis Plough, and J. Sook Chung.

